Our research is centered on studying the structure/function relationships of proteins. We use a variety of techniques to accomplish this goal and the research spans the fields of molecular biology, biochemistry and biophysics. Typically, proteins in bacterial systems are cloned and expressed, recombinant proteins are purified and characterized, conditions suitable for crystallization are identified, and then the structure of the crystallized protein is solved. The variety of techniques used provides plenty of opportunities for undergraduates to learn techniques that they will need in whatever post-baccalaureate path they choose.
SC-INBRE Research
Structure and Function Studies of Human Sphingosine Kinases 1 & 2
Lipid signaling molecules such as ceramide, sphingosine, and sphingosine-1-phosphate (S1P) have recently been shown to be increasingly important in determining cellular fate. The sphingosine kinase family of proteins helps regulate the balance of these molecules in the cell1 and one of the two family members, sphingosine kinase 1 (SK1) is overexpressed in various types of cancers. Increased expression of SK1 has been associated with tumor angiogenesis and resistance to radiation and chemotherapy.
At present, we have developed a homology model of sphingosine kinase 1 (See images below) and our collaborator within the department, Christian Grattan, has used the model to aid in the design of inhibitors of the protein. However, the sequence differences between the bacterial lipid kinases whose structures are known and were used to build the model and the human sphingosine kinases are significant. A crystallographic structure is essential toward developing compounds with maximum therapeutic effectiveness.
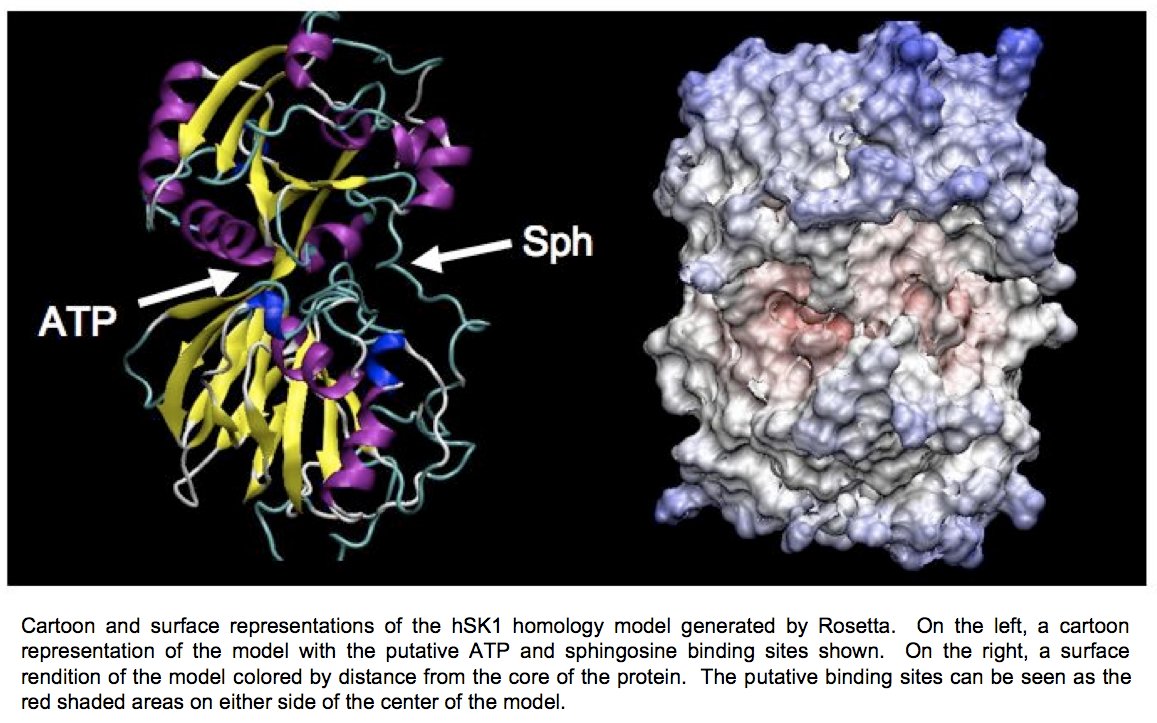
Determination of the three-dimensional structure of the proteins will allow for the crystallographic analysis of sphingosine kinases 1 and 2 in complex with substrates and biochemically verified inhibitors. Further determination of the structure of these enzymes in complex with sphingosine, ADP, and previously characterized inhibitors will allow for a better understanding of the reaction mechanism of this novel family of enzymes.
Other Research Projects
Crystallographic Analysis of Xylanase C from Bacillus subtilis
Biodegradation of lignocellulosic biomass for the purpose of bioconversion to fuel ethanol is receiving renewed attention spurred by the high cost of petroleum and the desire for energy independence. Emphasis has been directed toward optimizing physical and chemical disruption and improvement of the enzymatic pretreatment of this disrupted biomass to maximize the release of fermentable sugars. New hydrolytic enzyme activities that promise to increase the efficiency of this process are of great interest. Biochemical characterization of xylanases of glycosyl hydrolase family five (GH 5) have revealed a novel mode of action that may contribute to these efforts.
Two GH 5 xylanases have been purified and studied for their biochemical properties. XynA from the Gram-negative bacterium Erwinia chrysanthemi and XynC from the Gram-positive bacterium Bacillus subtilis share 40% amino acid identity. Of primary interest in both studies was the specificity and mode of action on the complex substrate glucuronoxylan and the characterization of the xylooligomeric products generated. Both XynA and XynC are glucuronoxylan specific and cleave the β-1,4 linked xylan main chain in an endo manner directed by the position of an α-1,2 linked glucuronate moiety. Results from several biochemical techniques all support the conclusion that cleavage occurs at the second β-1,4 linkage toward the reducing terminus from a glucuronate substituted xylose. This mode of action results in singly substituted glucuronoxylose oligosaccharide products with the glucuronic acid substitution penultimate to the reducing terminus (Figure 1). The novel specificity sets these enzymes apart from the more common endo-acting xylanases that cleave the xylan chain in assessable regions. This unique mode of action identifies the first endoxylanase family that degrades polymeric xylan in a directed rather than a random manner.
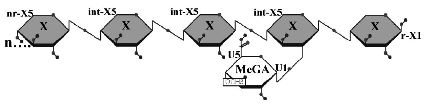
Limit product generated by the hydrolysis of glucuronoxylan by B. subtilis XynC. X, xylose; MeGA, 4-O-methylglucuronic acid; n, some number of beta-1,4-linked xylose residues.
XynC is the second GH 5 xylanase to be structurally studied. Recently, the high resolution crystallographic structure of XynC has been determined resulting from a collaborative effort involving the principal investigator, a student researcher, David Godwin, from Winthrop University and Dr. Franz St. John at the University of Maryland. Samples of recombinant protein were produced, purified and crystallized at Winthrop and then sent to the University of Maryland for crystallographic data collection. Diffraction data were collected on protein crystals using an x-ray source at the University of Maryland as well a portion of their time on a beamline at the Stanford Synchrotron Radiation Laboratory. These data were then analyzed by the principal investigator and the structure of the protein was solved to 1.8Å with molecular replacement techniques (Figure 2). The resulting structure has allowed for subsequent rapid determination of the structure of the enzyme in complex with a variety of oligoxylosides, oligomeric glucuronoxylosides and glucuronic acid moieties. Unfortunately, the electron densities of the oligosaccharides were unable to be resolved; most likely due to molecular motion and/or incomplete occupancy of the ligands in the active site. The structure of the enzyme in complex with glucuronic acid was able to be determined to 2.8Å with a clear density present for the pyranose ring. This structure provides direct evidence for the observed specificity of the enzyme and serves as the basis for the rationale of the current proposal.
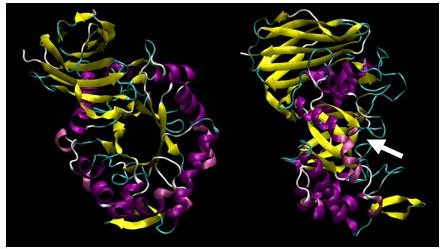
X-ray crystallographic structure of B. subtilis XynC at 1.8Å resolution. The model is colored such that alpha helices are purple and beta strands are colored yellow. The model on the right is a 90 degree rotation of the figure on the left. The active site cleft is indicated by the arrow.
Former Group Members
Rachel Glazener (2008): Earned her Master's Degree in Chemistry from the University of Tennessee and is teaching chemistry as an Adjunct Laboratory Instructor at Pellissippi State Community College in Knoxville, TN.
Kirk Godwin (2009): Working in the Fort Mill/Charlotte area.
Sabine Mahony (2009): Working in the San Antonio area.
Brad Angel (2009): Starting Veterinary School at the University of Georgia in Fall 2011.
Rachel Wilson (2010): Starting Pharmacy School at Presbyterian College in Fall 2011.
Cameron Waller (2011): Starting PhD research at the University of Utah in Fall 2011.
Zach Curry (2012): Starting MD/PhD program at Virginia Commonwealth University in Fall 2012.
Kennon Smith (2012): Working on his Ph.D. in Biochemistry at the University of Florida.
Tyler Couch (2013): Working in the Charlotte area.
Laura Wilt (2013): Working on her Ph.D. at the University of Georgia.
Harrison Toney (2014): Working in the Columbia area.
Current Group Members
Briana Murray (2015)