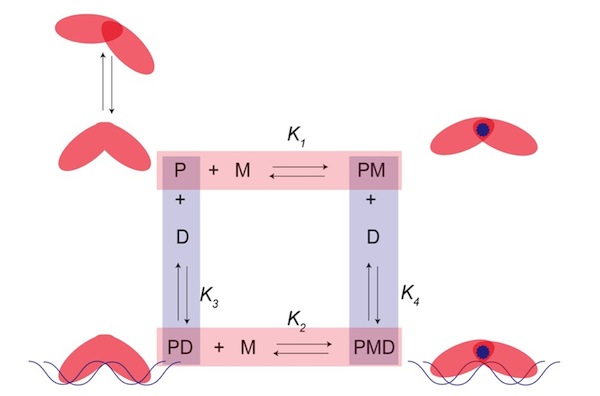
The energy that drives biological processes is the foundation of living systems. Every chemical process that occurs in or around an organism has an impact on how the cell allocates its metabolic resources. This principle forms the conceptual basis for regulatory proteins; non-homeostatic concentrations of a metabolite will induce an appropriate cellular response through allosteric communication between the effector and DNA binding sites. Fundamental thermodynamic information about processes such as these will enable more accurate predictions regarding the response elicited by a cell to specific stimuli. This entails a profound understanding of the general communication pathways that biological systems utilize. The research in my lab aims to develop and implement robust experimental methodologies capable of determining a complete thermodynamic description of these energies. Our approach, shown schematically in the figure just below, assigns each functional state of an allosteric regulatory protein to a corner of a closed thermodynamic cycle where P is the apoprotein, PM and PD are the protein-metal complex and protein-DNA complex, respectively, and PMD is the ternary complex that may form between the protein, metal and DNA. In this scheme, each side of the 'box' represents a chemical reaction that can occur in the cell. This allows us to relate the underlying thermodynamics of one side of the box to another and ultimately learn something about the energetics that allow the protein to carry out its function.
SC-INBRE Research
Cellular Copper Regulation
Mechanisms of copper homeostasis are of particular interest due to the need for organisms to devote significant cellular resources to regulate the concentration of this metal, which stems from the role copper plays in both essential and toxic processes. A large amount of information has been obtained about the biochemistry of copper. Similar to iron, copper's usefulness stems from a redox couple within the physiological range of reduction potentials (ε0 =0.153V), enabling electron transport through proteinacious copper centers. However, like iron, this redox couple also creates the possibility for deleterious effects such as the catalytic production of reactive oxidation species or inactivation of critical cellular enzymes through interactions with iron-sulfur clusters. It is therefore critical for cytosolic concentrations to be tightly regulated by the cell. Over the last decade, it has been repeatedly demonstrated that Cu+ is the physiologically relevant oxidation state for membrane transport, metallochaperone function and gene regulation. This complicates in vitro experiments, which have mainly focused on the Cu2+ oxidation state based on relative stabilities under typical laboratory conditions. Particularly, the presence of molecular oxygen will result in the spontaneous oxidation of Cu+ in a very favorable process (∆G˚ = -99 kcal mol-1), necessitating strict anaerobic conditions. Further, Cu+ will participate in a disproportion process (Equation 1).
2 Cu+ ⇌ Cu0(S) + Cu2+ (Equation 1)
This translates to a solution Cu+ concentration approximately 1000-fold below that of Cu2+. This, taken with the low solubility of many cuprous salts, makes Cu+ a challenging oxidation state to investigate. Fortunately, ligand selection can greatly influence the disproportionation equilibrium. The goal of this research project is to exploit this fact to stabilize Cu+, which can then be used for spectroscopic, potentiometric, and calorimetric titrations. These three complimentary techniques will paint a complete picture of Cu+ interactions with a number of interesting molecules and will allow us to take the next steps in determining the energetics of Cu+-specific allosteric regulatory proteins.
Additionally, we are investigating metal regulatory networks in Streptomyces coelicolor, an organism of interest due to the observation that metal homeostasis is directly linked to the biosynthesis of coelibactin, the product of a dedicated polyketide biosynthetic mechanism and precursor to multiple antibiotics. Notably, the genus Actinomyces, of which S. coelicolor is the most widely studied and understood, accounts for roughly half of the microbial antibiotics discovered to date. This project has two immediate goals:
- Characterize the metal and DNA binding properties of the Nickel Uptake Regulator (Nur)
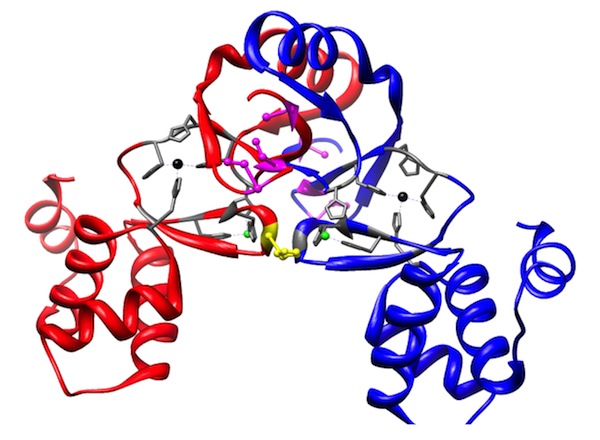
Using the crystal structure of Nur as a guide (below), we are working to biophysically characterize the metal binding and DNA binding chemistries of Nur. The goal of these experiments is to provide information about the allosteric coupling energies described above and mechanism understand the mechanism by which the metal binding site communicates with the DNA binding site.
- Investigate putative metal regulatory proteins.
Using the common families of metalloregulators as the basis for database searches, we have identified several candidate operons in the S. coelicolor A3(2) strain. The genes will be cloned into appropriate expression plasmids and transformed into Escherichia coli. The sensory metal for each of these candidate genes will be identified by Electrophoretic Mobility Shift Assays in the presence of DNA and various metal ions. The resulting pattern of band shifts will indicate which, if any, metals are able to induce a dissociation or association of the protein-DNA complex, which will directly guide future experiments. Of particular interest are proteins involved in sensing Zn2+ and Cu+. Once identified, these proteins will be purified to homogeneity, and thoroughly characterized biophysically.
Current Research Group
| Team Copper |
Team Streptomyces |
| Sharon Jenkins '11 |
Paisley Trantham '12 |
| Zayed Almadidy '12 |
Katie Bolling '12 |
| Destinee Johnson '14 |
Becca Toor '12 |